Projects
ConSReg
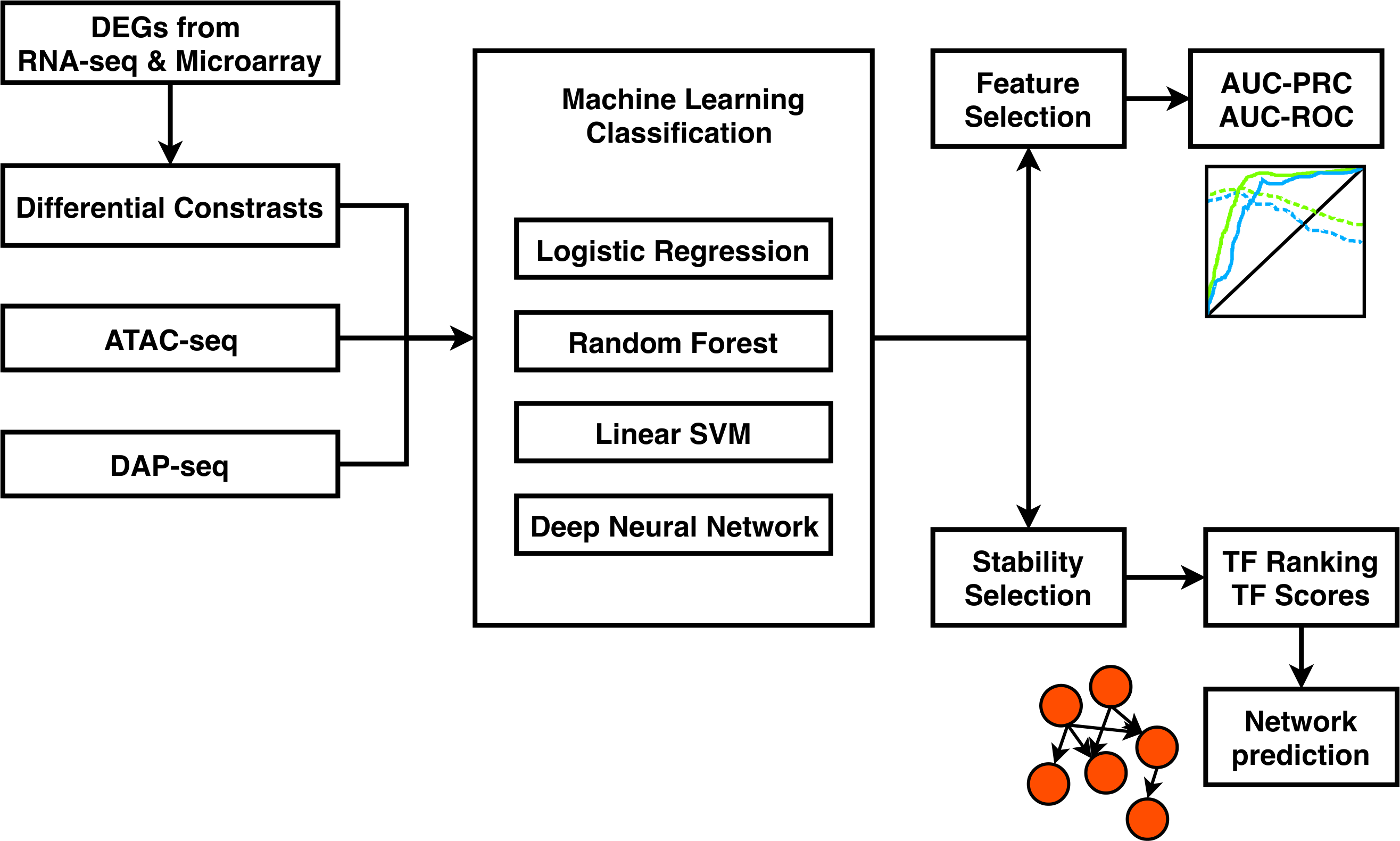 ConSReg stands for “Condition Specific Regulatory network inference engine”. ConSReg was designed to infer gene regulations using integrated heterogeneous genomic data (DAP-seq, ATAC-seq and single cell/bulk RNA-seq). ConSReg prioritizes key regulatory genes by using machine learning based regularization and stability selection (currently under review). ConSReg Python package is available at: https://github.com/LiLabAtVT/ConSReg
ConSReg stands for “Condition Specific Regulatory network inference engine”. ConSReg was designed to infer gene regulations using integrated heterogeneous genomic data (DAP-seq, ATAC-seq and single cell/bulk RNA-seq). ConSReg prioritizes key regulatory genes by using machine learning based regularization and stability selection (currently under review). ConSReg Python package is available at: https://github.com/LiLabAtVT/ConSReg
CoReg
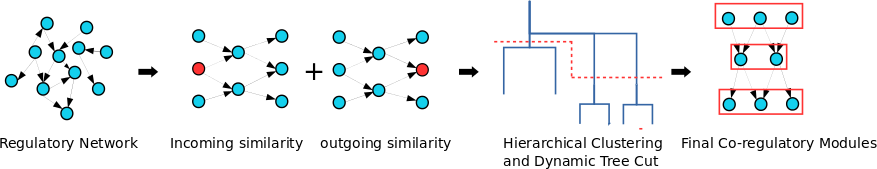 CoReg is a computational tool for identifying co-regulatory genes in large-scale networks. CoReg is provided as an R package, which can be applied in any organisms with genome-scale regulatory network data. You can find CoReg in my github repository The paper was recently published in BMC Systems Biology. CoReg R package is available at: https://github.com/LiLabAtVT/CoReg
CoReg is a computational tool for identifying co-regulatory genes in large-scale networks. CoReg is provided as an R package, which can be applied in any organisms with genome-scale regulatory network data. You can find CoReg in my github repository The paper was recently published in BMC Systems Biology. CoReg R package is available at: https://github.com/LiLabAtVT/CoReg
transEval
transEval is a computational tool for evaluating de novo transcriptome assembly result. This tool is currently under development and will be released soon.
